Applied
Q6. In this exercise, you will further analyze the Wage data set considered throughout this chapter.
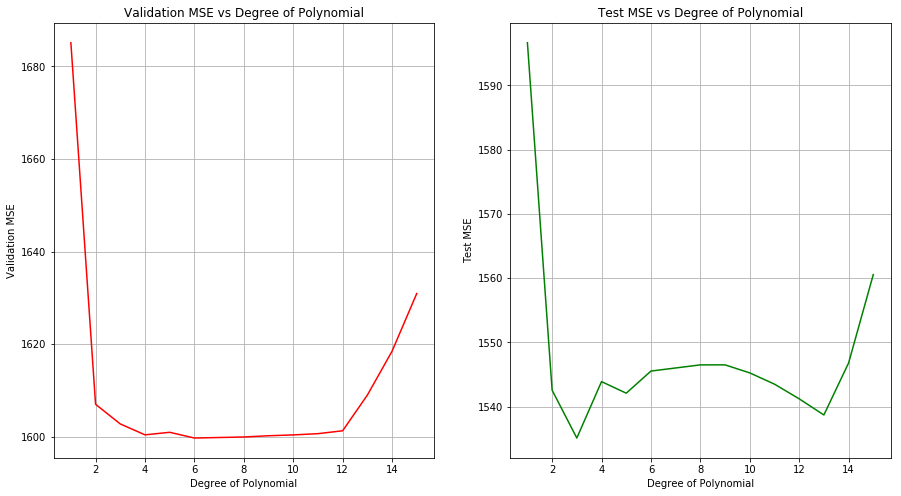
(a) Perform polynomial regression to predict wage using age. Use cross-validation to select the optimal degree d for the polynomial. What degree was chosen, and how does this compare to the results of hypothesis testing using ANOVA? Make a plot of the resulting polynomial fit to the data.
Sol: The optimal degree of polynomial selected from cross-validation is 4. From the ANOVA of models of degree 1 to 5, it is found that the models of degree 2 and 3 are significant. The test MSE shows the lowest value for degree 3 model as well.
import pandas as pd
import numpy as np
wage = pd.read_csv("data/Wage.csv")
wage.head()
| Unnamed: 0 | year | age | sex | maritl | race | education | region | jobclass | health | health_ins | logwage | wage | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 231655 | 2006 | 18 | 1. Male | 1. Never Married | 1. White | 1. < HS Grad | 2. Middle Atlantic | 1. Industrial | 1. <=Good | 2. No | 4.318063 | 75.043154 |
| 1 | 86582 | 2004 | 24 | 1. Male | 1. Never Married | 1. White | 4. College Grad | 2. Middle Atlantic | 2. Information | 2. >=Very Good | 2. No | 4.255273 | 70.476020 |
| 2 | 161300 | 2003 | 45 | 1. Male | 2. Married | 1. White | 3. Some College | 2. Middle Atlantic | 1. Industrial | 1. <=Good | 1. Yes | 4.875061 | 130.982177 |
| 3 | 155159 | 2003 | 43 | 1. Male | 2. Married | 3. Asian | 4. College Grad | 2. Middle Atlantic | 2. Information | 2. >=Very Good | 1. Yes | 5.041393 | 154.685293 |
| 4 | 11443 | 2005 | 50 | 1. Male | 4. Divorced | 1. White | 2. HS Grad | 2. Middle Atlantic | 2. Information | 1. <=Good | 1. Yes | 4.318063 | 75.043154 |
from sklearn import preprocessing
from sklearn.model_selection import LeaveOneOut
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
np.random.seed(5)
def polynomial_regression(X_train, Y_train, X_test, Y_test, M):
validation_MSE = {}
test_MSE = {}
for m in M:
poly = preprocessing.PolynomialFeatures(degree=m)
X_ = poly.fit_transform(X_train)
mse = 0
loo = LeaveOneOut() # Leave one out cross-validation
for train_index, test_index in loo.split(X_):
X, X_CV = X_[train_index], X_[test_index]
Y, Y_CV = Y_train[train_index], Y_train[test_index]
# Linear Regression (including higher order predictors)
model = LinearRegression(fit_intercept=True)
model.fit(X, Y)
p = model.predict(X_CV)
mse += mean_squared_error(p, Y_CV)
validation_MSE[m] = mse/len(X_)
# Compute test MSE for the model
model = LinearRegression(fit_intercept=True)
model.fit(X_, Y_train)
p = model.predict(poly.fit_transform(X_test))
test_MSE[m] = mean_squared_error(p, Y_test)
# Plot validation MSE
lists = sorted(validation_MSE.items()) # sorted by key, return a list of tuples
x, y = zip(*lists) # unpack a list of pairs into two tuples
fig = plt.figure(figsize=(15, 8))
ax = fig.add_subplot(121)
plt.plot(x, y, color='r')
plt.grid()
ax.set_xlabel('Degree of Polynomial')
ax.set_ylabel('Validation MSE')
ax.set_title('Validation MSE vs Degree of Polynomial')
lists = sorted(test_MSE.items()) # sorted by key, return a list of tuples
x, y = zip(*lists) # unpack a list of pairs into two tuples
ax = fig.add_subplot(122)
plt.plot(x, y, color='g')
plt.grid()
ax.set_xlabel('Degree of Polynomial')
ax.set_ylabel('Test MSE')
ax.set_title('Test MSE vs Degree of Polynomial')
plt.show()
X_train, X_test, y_train, y_test = train_test_split(wage[['age']], wage[['wage']], test_size=0.1)
polynomial_regression(X_train, y_train.values, X_test, y_test, [1,2,3,4,5,6,8,9,10,11,12,13,14,15])
import statsmodels.api as sm
poly = preprocessing.PolynomialFeatures(degree=1)
X_ = poly.fit_transform(wage[['age']])
model1 = sm.OLS(wage[['wage']], X_)
model1 = model1.fit()
poly = preprocessing.PolynomialFeatures(degree=2)
X_ = poly.fit_transform(wage[['age']])
model2 = sm.OLS(wage[['wage']], X_)
model2 = model2.fit()
poly = preprocessing.PolynomialFeatures(degree=3)
X_ = poly.fit_transform(wage[['age']])
model3 = sm.OLS(wage[['wage']], X_)
model3 = model3.fit()
poly = preprocessing.PolynomialFeatures(degree=4)
X_ = poly.fit_transform(wage[['age']])
model4 = sm.OLS(wage[['wage']], X_)
model4 = model4.fit()
poly = preprocessing.PolynomialFeatures(degree=5)
X_ = poly.fit_transform(wage[['age']])
model5 = sm.OLS(wage[['wage']], X_)
model5 = model5.fit()
sm.stats.anova_lm(model1, model2, model3, model4, model5)
/Users/amitrajan/Desktop/PythonVirtualEnv/Python3_VirtualEnv/lib/python3.6/site-packages/scipy/stats/_distn_infrastructure.py:879: RuntimeWarning: invalid value encountered in greater
return (self.a < x) & (x < self.b)
/Users/amitrajan/Desktop/PythonVirtualEnv/Python3_VirtualEnv/lib/python3.6/site-packages/scipy/stats/_distn_infrastructure.py:879: RuntimeWarning: invalid value encountered in less
return (self.a < x) & (x < self.b)
/Users/amitrajan/Desktop/PythonVirtualEnv/Python3_VirtualEnv/lib/python3.6/site-packages/scipy/stats/_distn_infrastructure.py:1821: RuntimeWarning: invalid value encountered in less_equal
cond2 = cond0 & (x <= self.a)
| df_resid | ssr | df_diff | ss_diff | F | Pr(>F) | |
|---|---|---|---|---|---|---|
| 0 | 2998.0 | 5.022216e+06 | 0.0 | NaN | NaN | NaN |
| 1 | 2997.0 | 4.793430e+06 | 1.0 | 228786.010128 | 143.593107 | 2.363850e-32 |
| 2 | 2996.0 | 4.777674e+06 | 1.0 | 15755.693664 | 9.888756 | 1.679202e-03 |
| 3 | 2995.0 | 4.771604e+06 | 1.0 | 6070.152124 | 3.809813 | 5.104620e-02 |
| 4 | 2994.0 | 4.770322e+06 | 1.0 | 1282.563017 | 0.804976 | 3.696820e-01 |
(b) Fit a step function to predict wage using age, and perform crossvalidation to choose the optimal number of cuts. Make a plot of the fit obtained. Code Source: https://rpubs.com/ppaquay/65563
Q9. This question uses the variables dis (the weighted mean of distances to five Boston employment centers) and nox (nitrogen oxides concentration in parts per 10 million) from the Boston data. We will treat dis as the predictor and nox as the response.
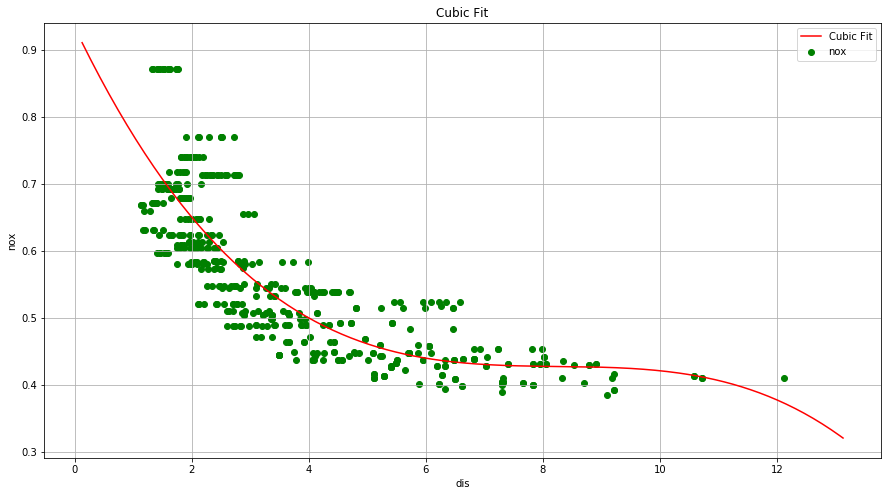
(a) Use the poly() function to fit a cubic polynomial regression to predict nox using dis. Report the regression output, and plot the resulting data and polynomial fits.
boston = pd.read_csv("data/Boston.csv")
boston.dropna(inplace=True)
poly = preprocessing.PolynomialFeatures(degree=3)
X_ = poly.fit_transform(boston[['dis']])
model = sm.OLS(boston[['nox']], X_)
result = model.fit()
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: nox R-squared: 0.715
Model: OLS Adj. R-squared: 0.713
Method: Least Squares F-statistic: 419.3
Date: Thu, 20 Sep 2018 Prob (F-statistic): 2.71e-136
Time: 19:22:18 Log-Likelihood: 690.44
No. Observations: 506 AIC: -1373.
Df Residuals: 502 BIC: -1356.
Df Model: 3
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 0.9341 0.021 45.110 0.000 0.893 0.975
x1 -0.1821 0.015 -12.389 0.000 -0.211 -0.153
x2 0.0219 0.003 7.476 0.000 0.016 0.028
x3 -0.0009 0.000 -5.124 0.000 -0.001 -0.001
==============================================================================
Omnibus: 64.176 Durbin-Watson: 0.286
Prob(Omnibus): 0.000 Jarque-Bera (JB): 87.386
Skew: 0.917 Prob(JB): 1.06e-19
Kurtosis: 3.886 Cond. No. 2.10e+03
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 2.1e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
x = np.linspace(boston['dis'].min()-1, boston['dis'].max()+1, 256, endpoint = True)
y = result.params.const + result.params.x1*x + result.params.x2*(x**2) + result.params.x3*(x**3)
fig = plt.figure(figsize=(15, 8))
plt.plot(x, y, 'red', label="Cubic Fit")
plt.scatter(boston['dis'], boston['nox'], alpha=1, color='green')
plt.xlabel('dis')
plt.ylabel('nox')
plt.title('Cubic Fit')
plt.legend(loc='best')
plt.grid()
plt.show()
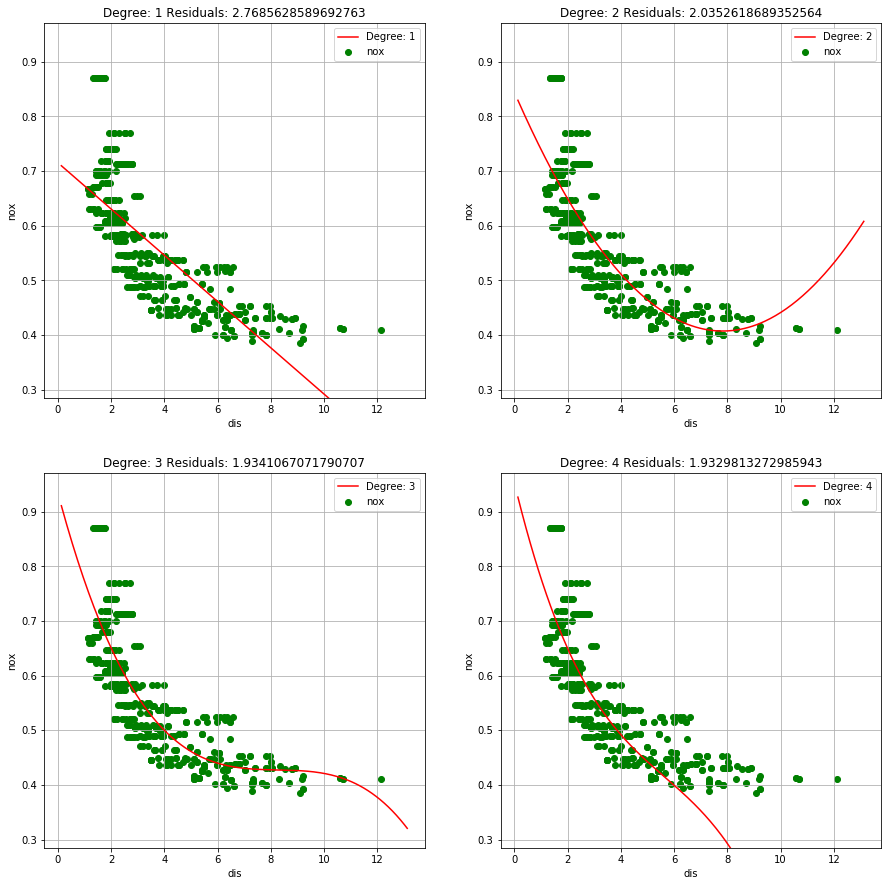
(b) Plot the polynomial fits for a range of different polynomial degrees (say, from 1 to 10), and report the associated residual sum of squares.
def add_polt(ax, x, y, RSS, degree):
ax.plot(x, y, 'red', label="Degree: " +str(degree))
ax.scatter(boston['dis'], boston['nox'], alpha=1, color='green')
ax.set_ylim(boston['nox'].min()-0.1, boston['nox'].max()+0.1)
ax.set_xlabel('dis')
ax.set_ylabel('nox')
ax.set_title('Degree: ' +str(degree) + ' Residuals: ' + str(RSS))
ax.legend(loc='best')
ax.grid()
fig = plt.figure(figsize=(15, 40))
x = np.linspace(boston['dis'].min()-1, boston['dis'].max()+1, 256, endpoint = True)
degree = 1
poly = preprocessing.PolynomialFeatures(degree=degree)
X_ = poly.fit_transform(boston[['dis']])
model = sm.OLS(boston[['nox']], X_)
result = model.fit()
y = result.params.const + result.params.x1*x
ax = fig.add_subplot(5, 2, degree)
add_polt(ax, x, y, result.ssr, degree)
degree = 2
poly = preprocessing.PolynomialFeatures(degree=degree)
X_ = poly.fit_transform(boston[['dis']])
model = sm.OLS(boston[['nox']], X_)
result = model.fit()
y = result.params.const + result.params.x1*x + result.params.x2*(x**2)
ax = fig.add_subplot(5, 2, degree)
add_polt(ax, x, y, result.ssr, degree)
degree = 3
poly = preprocessing.PolynomialFeatures(degree=degree)
X_ = poly.fit_transform(boston[['dis']])
model = sm.OLS(boston[['nox']], X_)
result = model.fit()
y = result.params.const + result.params.x1*x + result.params.x2*(x**2) + result.params.x3*(x**3)
ax = fig.add_subplot(5, 2, degree)
add_polt(ax, x, y, result.ssr, degree)
degree = 4
poly = preprocessing.PolynomialFeatures(degree=degree)
X_ = poly.fit_transform(boston[['dis']])
model = sm.OLS(boston[['nox']], X_)
result = model.fit()
y = result.params.const + result.params.x1*x + result.params.x2*(x**2) + result.params.x3*(x**3)
+ result.params.x4*(x**4)
ax = fig.add_subplot(5, 2, degree)
add_polt(ax, x, y, result.ssr, degree)
plt.show()
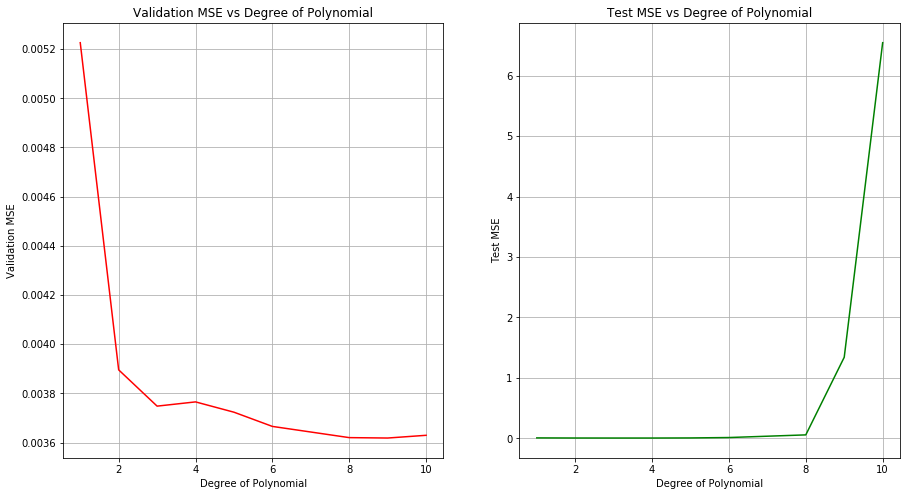
(c) Perform cross-validation or another approach to select the optimal degree for the polynomial, and explain your results.
Sol: The optimal degree of polynomial is 8. Achieved test MSE at this value is decent as well.
np.random.seed(5)
X_train, X_test, y_train, y_test = train_test_split(boston[['dis']], boston[['nox']], test_size=0.2)
polynomial_regression(X_train, y_train.values, X_test, y_test, [1,2,3,4,5,6,8,9,10])
(d) Use the bs() function to fit a regression spline to predict nox using dis. Report the output for the fit using four degrees of freedom. How did you choose the knots? Plot the resulting fit.
(e) Now fit a regression spline for a range of degrees of freedom, and plot the resulting fits and report the resulting RSS. Describe the results obtained.
(f) Perform cross-validation or another approach in order to select the best degrees of freedom for a regression spline on this data. Describe your results.
Q10. This question relates to the College data set.
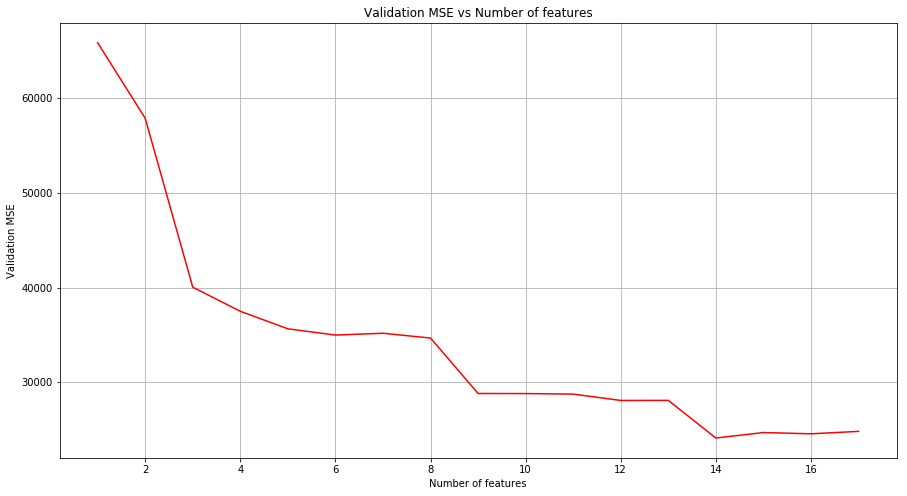
(a) Split the data into a training set and a test set. Using out-of-state-tuition as the response and the other variables as the predictors, perform forward stepwise selection on the training set in order to identify a satisfactory model that uses just a subset of the predictors.
college = pd.read_csv("data/College.csv")
college = college.rename(columns={'Unnamed: 0': 'Name'})
college['Private'] = college['Private'].map({'Yes': 1, 'No': 0})
college.head()
| Name | Private | Apps | Accept | Enroll | Top10perc | Top25perc | F.Undergrad | P.Undergrad | Outstate | Room.Board | Books | Personal | PhD | Terminal | S.F.Ratio | perc.alumni | Expend | Grad.Rate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Abilene Christian University | 1 | 1660 | 1232 | 721 | 23 | 52 | 2885 | 537 | 7440 | 3300 | 450 | 2200 | 70 | 78 | 18.1 | 12 | 7041 | 60 |
| 1 | Adelphi University | 1 | 2186 | 1924 | 512 | 16 | 29 | 2683 | 1227 | 12280 | 6450 | 750 | 1500 | 29 | 30 | 12.2 | 16 | 10527 | 56 |
| 2 | Adrian College | 1 | 1428 | 1097 | 336 | 22 | 50 | 1036 | 99 | 11250 | 3750 | 400 | 1165 | 53 | 66 | 12.9 | 30 | 8735 | 54 |
| 3 | Agnes Scott College | 1 | 417 | 349 | 137 | 60 | 89 | 510 | 63 | 12960 | 5450 | 450 | 875 | 92 | 97 | 7.7 | 37 | 19016 | 59 |
| 4 | Alaska Pacific University | 1 | 193 | 146 | 55 | 16 | 44 | 249 | 869 | 7560 | 4120 | 800 | 1500 | 76 | 72 | 11.9 | 2 | 10922 | 15 |
from sklearn.linear_model import LinearRegression
from sklearn.feature_selection import RFE
from sklearn.metrics import mean_squared_error
np.random.seed(1)
msk = np.random.rand(len(college)) < 0.8
train = college[msk]
test = college[~msk]
total_features = 17
validation_MSE = {}
for m in range(1, total_features+1):
estimator = LinearRegression(fit_intercept=True)
selector = RFE(estimator, m, step=1)
selector.fit(train.drop(['Name', 'Outstate'], axis=1), train['Outstate'])
predictions = selector.predict(test.drop(['Name', 'Outstate'], axis=1))
mse = mean_squared_error(predictions, test['Outstate'])
validation_MSE[m] = mse/len(test)
# Plot validation MSE
lists = sorted(validation_MSE.items()) # sorted by key, return a list of tuples
x, y = zip(*lists) # unpack a list of pairs into two tuples
fig = plt.figure(figsize=(15, 8))
ax = fig.add_subplot(111)
plt.plot(x, y, color='r')
plt.grid()
ax.set_xlabel('Number of features')
ax.set_ylabel('Validation MSE')
ax.set_title('Validation MSE vs Number of features')
Text(0.5,1,'Validation MSE vs Number of features')
(b) Fit a GAM on the training data, using out-of-state tuition as the response and the features selected in the previous step as the predictors. Plot the results, and explain your findings.
(c) Evaluate the model obtained on the test set, and explain the results obtained.
(d) For which variables, if any, is there evidence of a non-linear relationship with the response?